一键计算泛癌中基因表达与肿瘤干性指数的关系
看教程不够直观,那就看视频吧! >>点击加载视频
众所周知,现阶段研究肿瘤基本离不开TCGA,研究单个基因的话更离不开单个基因在泛癌中的表达,当然我们也可以分析 肿瘤细胞干性与基因表达的关系,我们从先前的研究中(Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation)获取了通过mRNA表达和甲基化signature计算而来的六种肿瘤干性指数,如下:
The RNA based Stemness Scores derived by the Stemness group.
RNAss: RNA expression-based (All set of available genes) this score will drive the main figures in PancanAtlas paper.
EREG.EXPss: Epigenetically regulated RNA expression-based (103 genes).
The DNA methylation based Stemness Scores derived by the Stemness group.
DNAss: DNA methylation-based (Stem cell signature probes (219 probes), that combines the 3 signatures listed below). This score will drive the main figures in the PancanAtlas paper
EREG-METHss: Epigenetically regulated DNA methylation-based (87 probes).
DMPss: Differentially methylated probes-based (62 probes)
ENHss: Enhancer Elements/DNAmethylation-based (82 probes)
在这里我们介绍一个傻瓜式的工具助你高效的分析,不用输入数据,鼠标点点点即可,并且支持动态调整,此外它还提供了结直肠癌、胶质瘤、混合肾癌、胃和食管癌等合并肿瘤的基因表达与六种肿瘤干性指数的相关性分析,这在其他工具中是没有的
 输入一个基因就可以了,剩下的默认即可,会得到图片如下,图片的详细调整方式可以 参考 这里:
输入一个基因就可以了,剩下的默认即可,会得到图片如下,图片的详细调整方式可以 参考 这里:

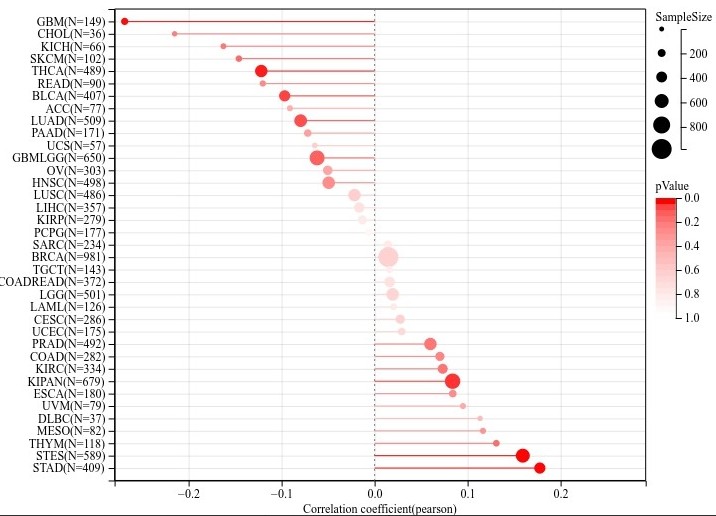
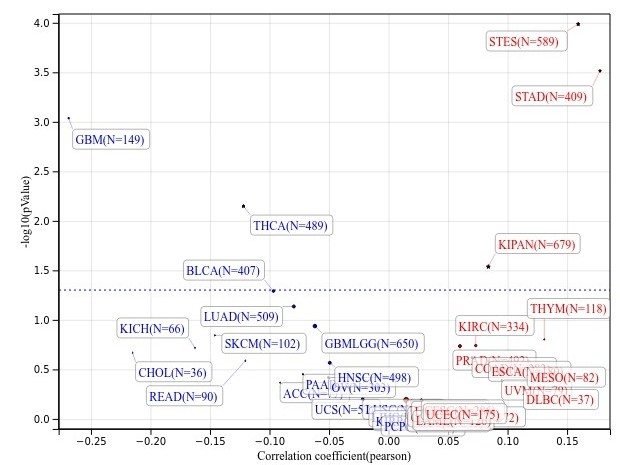
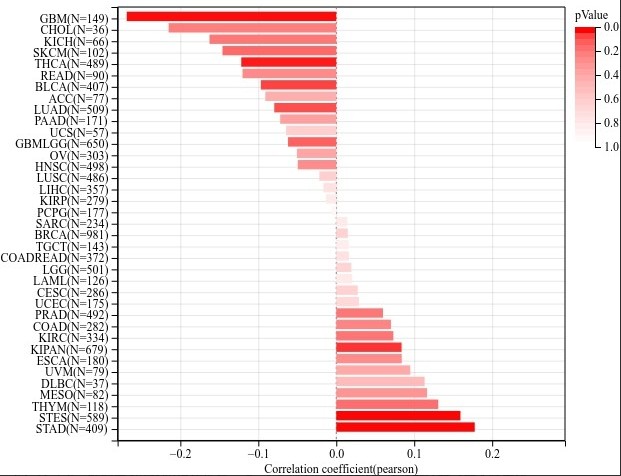
文章写作示例在网页下方:
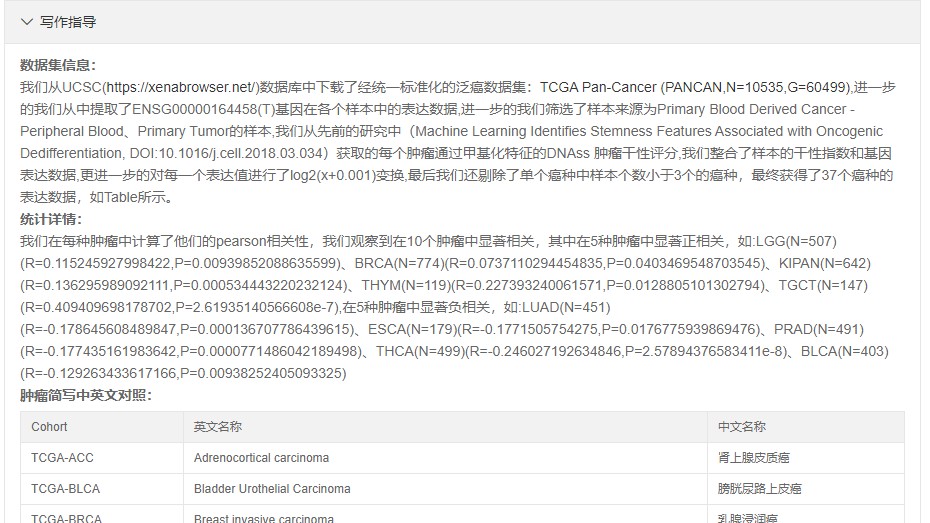
- 发表于 2021-10-07 12:06
- 阅读 ( 10642 )
- 分类:软件工具
