你把下半部分的基因排在前面再画图应该就可以了吧
10 关于pheatmap画热图的问题
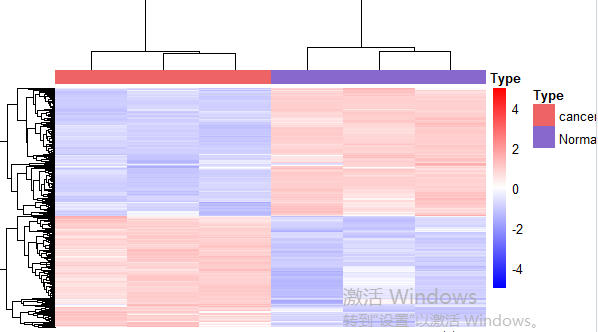 请问各位老师,怎么改变基因顺序,即让各组的红蓝色对调一下,让cancer组红色在上蓝色在下,normal组红色在下蓝色在上?下面是我的代码,请问该如何调整
请问各位老师,怎么改变基因顺序,即让各组的红蓝色对调一下,让cancer组红色在上蓝色在下,normal组红色在下蓝色在上?下面是我的代码,请问该如何调整
lncRNA_exp_heatmap<-lncRNA_exp[row.names(final_DElncRNA),]
ann_col = data.frame(Type = factor(rep(c("cancer","Normal"),c(3,3)) ))
rownames(ann_col) <- colnames(lncRNA_exp_heatmap)
ann_colors = list(Type = c(cancer = "indianred2", Normal = "mediumpurple3"))
bk = unique(c(seq(-5,5, length=1000)))
pheatmap(lncRNA_exp_heatmap,
annotation_col = ann_col,annotation_legend=TRUE,annotation_colors = ann_colors,breaks = bk,scale = 'row',
cluster_rows=TRUE,cluster_cols=TRUE,show_colnames = TRUE,show_rownames = FALSE,
color = colorRampPalette(c("blue","white","red"))(1000))
请先 登录 后评论
2 个回答
- 1 关注
- 0 收藏,4648 浏览
- 肖晓 提出于 2020-07-02 20:36