复现了你的结果:
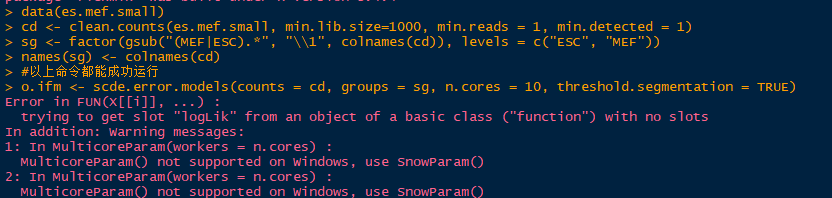
看提示应该是多核运算不支持的问题
source("https://bioconductor.org/biocLite.R")
library(scde) data(es.mef.small) cd <- clean.counts(es.mef.small, min.lib.size=1000, min.reads = 1, min.detected = 1) sg <- factor(gsub("(MEF|ESC).*", "\\1", colnames(cd)), levels = c("ESC", "MEF")) names(sg) <- colnames(cd) #以上命令都能成功运行 o.ifm <- scde.error.models(counts = cd, groups = sg, n.cores = 10, threshold.segmentation = TRUE) #运行此命令时报错如下:
Error in FUN(X[[i]], ...) :
trying to get slot "logLik" from an object of a basic class ("function") with no slots
自我检查:
1,R版本和SCDE包不冲突,library正常,所依赖的包都有
2,以为是命令引起程序误会,使用scde::scde.error.models 命令也不可以
请求大神赐教