Deseq2出现的错误和我运行出现的一样,咨询国祝老师,说正在调试软件。
EdgeR的结果若不一样,那这个软件可靠性有待考查啊!
下载了TCGA的Rna-seq数据后分别用了DECenter和r中的edgeR包进行分析,完全按照教程来操作的,但得出结果有很大不同,求各位大神解答!还有按提示方法用DESeq2包进行分析时出现错误,求解!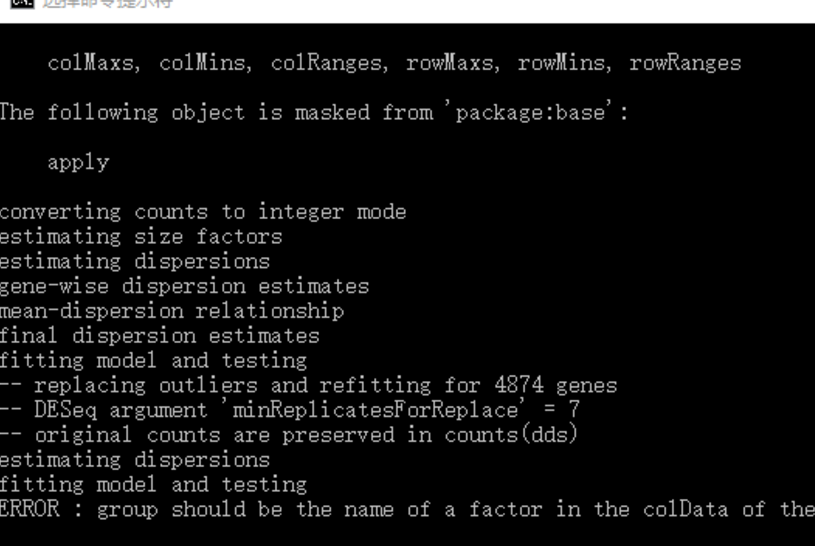

edgeR代码如下:
#edgeR
source("http://bioconductor.org/biocLite.R")
biocLite("edgeR")
install.packages("gplots")
foldChange=2
padj=0.01
setwd("F:\\2database\\colorectal\\mrna") #设置工作目录
library("edgeR")
rt=read.table("mrna.txt",sep="\t",header=T,check.names=F) #改成自己的文件名
rt=as.matrix(rt)
rownames(rt)=rt[,1]
exp=rt[,2:ncol(rt)]
dimnames=list(rownames(exp),colnames(exp))
data=matrix(as.numeric(as.matrix(exp)),nrow=nrow(exp),dimnames=dimnames)
data=avereps(data)
data=data[rowMeans(data)>1,]
group=c(rep("normal",375),rep("tumor",18)) #按照癌症和正常样品数目修改
design <- model.matrix(~group)
y <- DGEList(counts=data,group=group)
y <- calcNormFactors(y)
y <- estimateCommonDisp(y)
y <- estimateTagwiseDisp(y)
et <- exactTest(y,pair = c("normal","tumor"))
topTags(et)
ordered_tags <- topTags(et, n=100000)
allDiff=ordered_tags$table
allDiff=allDiff[is.na(allDiff$FDR)==FALSE,]
diff=allDiff
newData=y$pseudo.counts
write.table(diff,file="edgerOut.xls",sep="\t",quote=F)
diffSig = diff[(diff$FDR < padj & (diff$logFC>foldChange | diff$logFC<(-foldChange))),]
write.table(diffSig, file="diffSig.xls",sep="\t",quote=F)
diffUp = diff[(diff$FDR < padj & (diff$logFC>foldChange)),]
write.table(diffUp, file="up.xls",sep="\t",quote=F)
diffDown = diff[(diff$FDR < padj & (diff$logFC<(-foldChange))),]
write.table(diffDown, file="down.xls",sep="\t",quote=F)
normalizeExp=rbind(id=colnames(newData),newData)
write.table(normalizeExp,file="normalizeExp.txt",sep="\t",quote=F,col.names=F) #输出所有基因校正后的表达值(normalizeExp.txt)
diffExp=rbind(id=colnames(newData),newData[rownames(diffSig),])
write.table(diffExp,file="diffmRNAExp.txt",sep="\t",quote=F,col.names=F) #输出差异基因校正后的表达值(diffmRNAExp.txt)