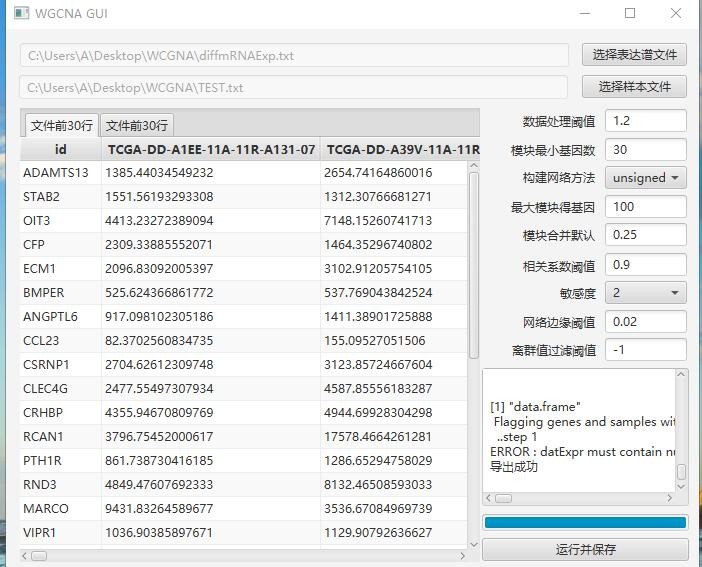
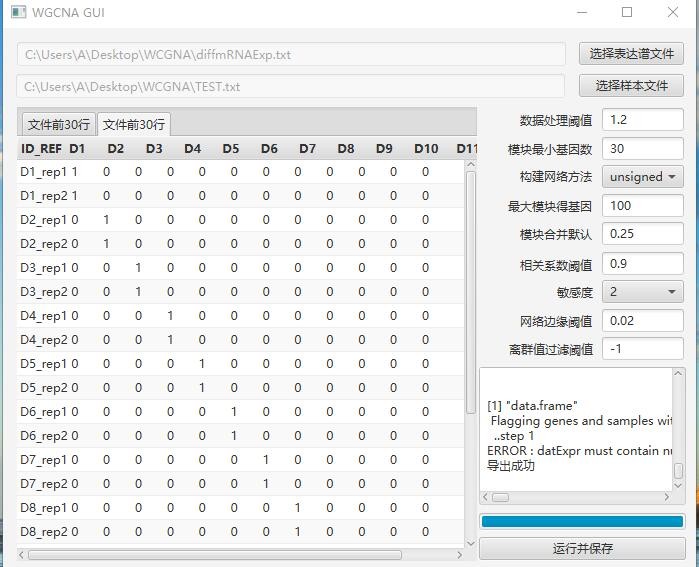
从报错来看是表达矩阵的数据类型不对,你重新保存一份数据看看,用execel另存为文本文件(Tab分割)
10 sanger box 做WGCNA,为什么不成功,哪里出了问题?
运行结果如下,图一和图二是软件界面,图三是结果界面
读取文件完毕
开始运行
文本处理完毕
C:\R\R-3.3.3\bin\Rscript.exe
C:\Users\A\Desktop\Sanger_V1.0.8\source\softs\2018-08-24-09-12-06-5b7f5b666949f\WGCNA_Pipline.R
开始运行R
"C:\R\R-3.3.3\bin\Rscript.exe" "C:\Users\A\Desktop\Sanger_V1.0.8\source\softs\2018-08-24-09-12-06-5b7f5b666949f\WGCNA_Pipline.R" "C:\Users\A\Desktop\WCGNA" "success_diffmRNAExp.txt" "success_TEST.txt" "1.2" "30" "unsigned" "100" "0.25" "0.9" "2" "0.02" "-1" "1"
==========================================================================
*
* Package WGCNA 1.63 loaded.
*
* Important note: It appears that your system supports multi-threading,
* but it is not enabled within WGCNA in R.
* To allow multi-threading within WGCNA with all available cores, use
*
* allowWGCNAThreads()
*
* within R. Use disableWGCNAThreads() to disable threading if necessary.
* Alternatively, set the following environment variable on your system:
*
* ALLOW_WGCNA_THREADS=
*
* for example
*
* ALLOW_WGCNA_THREADS=8
*
* To set the environment variable in linux bash shell, type
*
* export ALLOW_WGCNA_THREADS=8
*
* before running R. Other operating systems or shells will
* have a similar command to achieve the same aim.
*
==========================================================================
[1] "data.frame"
Flagging genes and samples with too many missing values...
..step 1
ERROR : datExpr must contain numeric data.
导出成功
- 2 关注
- 0 收藏,7618 浏览
- 提出于 2018-11-01 09:38